Our research broadly focuses on host-microbe symbioses, evolutionary genomics, and bioinformatics; with special reference to the emergence, evolution, and genotypic/phenotypic consequences of new symbiotic associations (🦠 ↔ 🪱,🐜,…) for both host and symbionts. Two main axes drive our current research:
Obligate nutritional endosymbionts
Read more
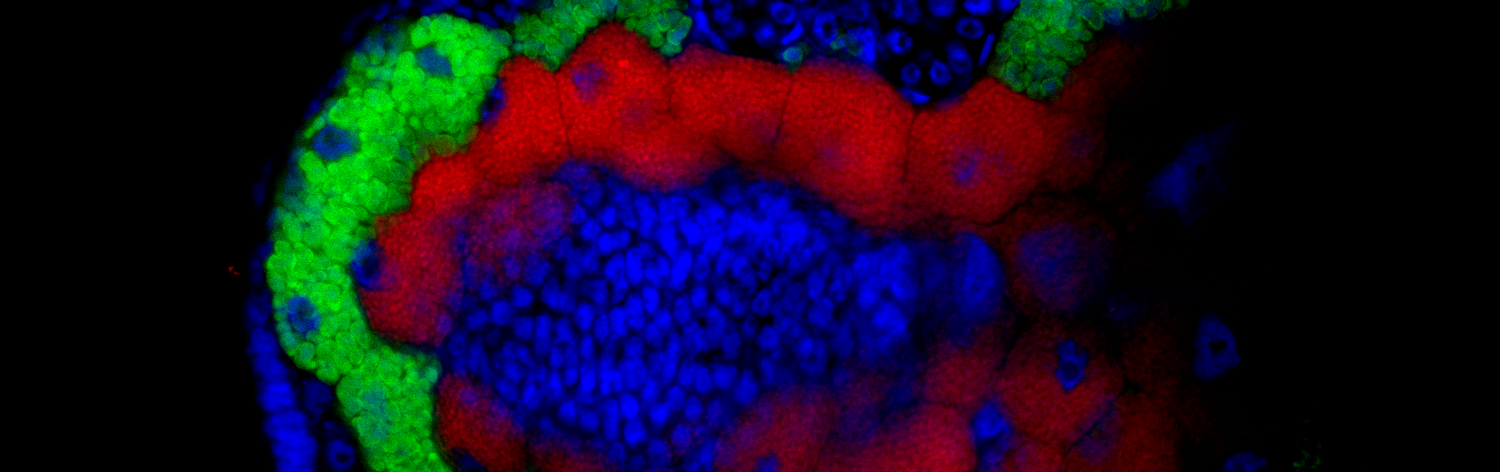
Animals with nutrient restricted diets, such as plant phloem (plant pests) or mammalian blood (animal parasites and disease vectors), lack essential nutrients from their diets, namely essential amino acids and B vitamins. They depend mainly on obligate symbiotic bacteria to complement their diets. These symbiotic bacteria become host-restricted and vertically-transmitted, often evolving small compact genomes with a limited genetic repertoire specialised in synthesising essential nutrients for the host. Sometimes, these symbionts lose key pathways for the production of these compounds, which can lead to symbiont replacement or to the acquisition of a secondary co-obligate symbiont (symbiont complementation). We are interested in the processes that underlay these two phenomena. We use whole genome sequencing of mainly the symbionts, but also the hosts, to infer the possible interactions that exist among the co-existing symbionts as well as between host and symbionts. Additionally, through the use microscopy, we link changes in the genome and nutritional interactions to the symbiont’s phenotypic changes (e.g. cell shape and tissue tropism). The two main systems we work on are aphids and strict blood-feeding leeches, but also on other critters through collaborations.
Assembly and maintenance of gut-associated symbionts
Read more
Symbiotic bacteria not only inhabit bacteriomes, but can also be found residing in different host tissues. Some of these are essential to the correct development of their hosts, protect them, and/or aid in the digestion of compounds, among other things. While the complexity of communities of host-associated bacteria found in the gut of most vertebrates limits the close analysis of the roles and contributions of individual species, the simple and often defined communities found in some invertebrates allow such investigations. The gut-associated microbiota of many invertebrates with a nutrient-restricted diet display such simplicity: few dominant strains and often a well-defined localisation. Currently, we are investigating the gut-associated bacteria from aphids, as well as the inhabitants of the different organs across the leech’s digestive system. We approach the study of these organisms using microbial culturing, genome sequencing (host and symbionts), microscopy, and phylogenetics.
📱 Social media
 BlueSky
BlueSky
- # Fe️eds: #S️ymbioSky
- 📦️ Starter packs: Microbial Symbiosis
💸 Funders


 Biology and Evolution of Symbiosis (BESymb)
Biology and Evolution of Symbiosis (BESymb)