-
Welcome Pegah!

Last week, Pegah Kalatehjari joined the team! Pegah joins us from Iran, with a background in aquaculture, leech biology and reproduction. She will work on the...
-
Back from ESEB 2025!

ESEB 2025 just wrapped up last week. It was a great week of talks, posters, and informal chats about all things evolution. Particularly, it was great...
-
Visit to Shenyang Agricultural University

Taking advantage of the trip to China for SMBE 2025, Alejandro visited the group of Jun-Bo Luan (https://www.junboluanlab.com/) at the Shenyang Agricultural University (沈阳农业大学). It was...
-
Symbiosis symposium @SMBE 2025!

Along with Jun-Bo Luan (https://www.junboluanlab.com/), we organised the Symbiosis symposium at SMBE 2025 (Beijing, China). We were thrilled to see the turnout and the great talks...
-
New work out! New protist genomes, codon usage, and giant viruses!
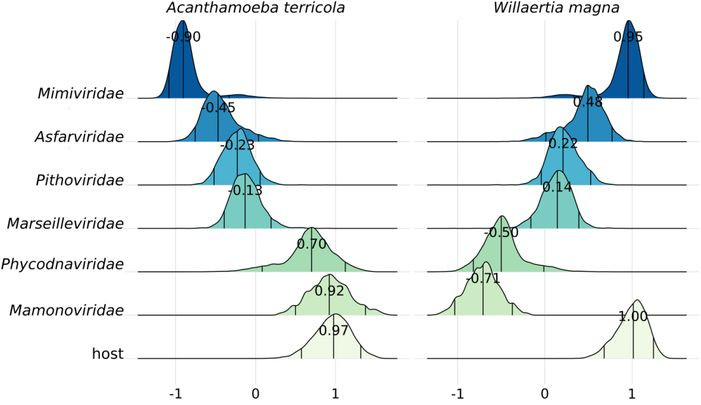
In collaboration with Anouk Willemsen (Uniersity of Vienna), we sequenced high-quality genomes of protists known as hosts of giant viruses (GVs). Matching of codon usage preferences...
-
Project funded! “SymBirth: Development of an artificial symbiont’s birth” (FWF 1000 ideas)

Very happy the first group project SymBirth: Development of an artificial symbiont’s birth has been funded by the FWF’s 1000 ideas program. This program aims to...
-
New work out! Discordance between phylogenomic datasets in aphids: who is telling the truth?
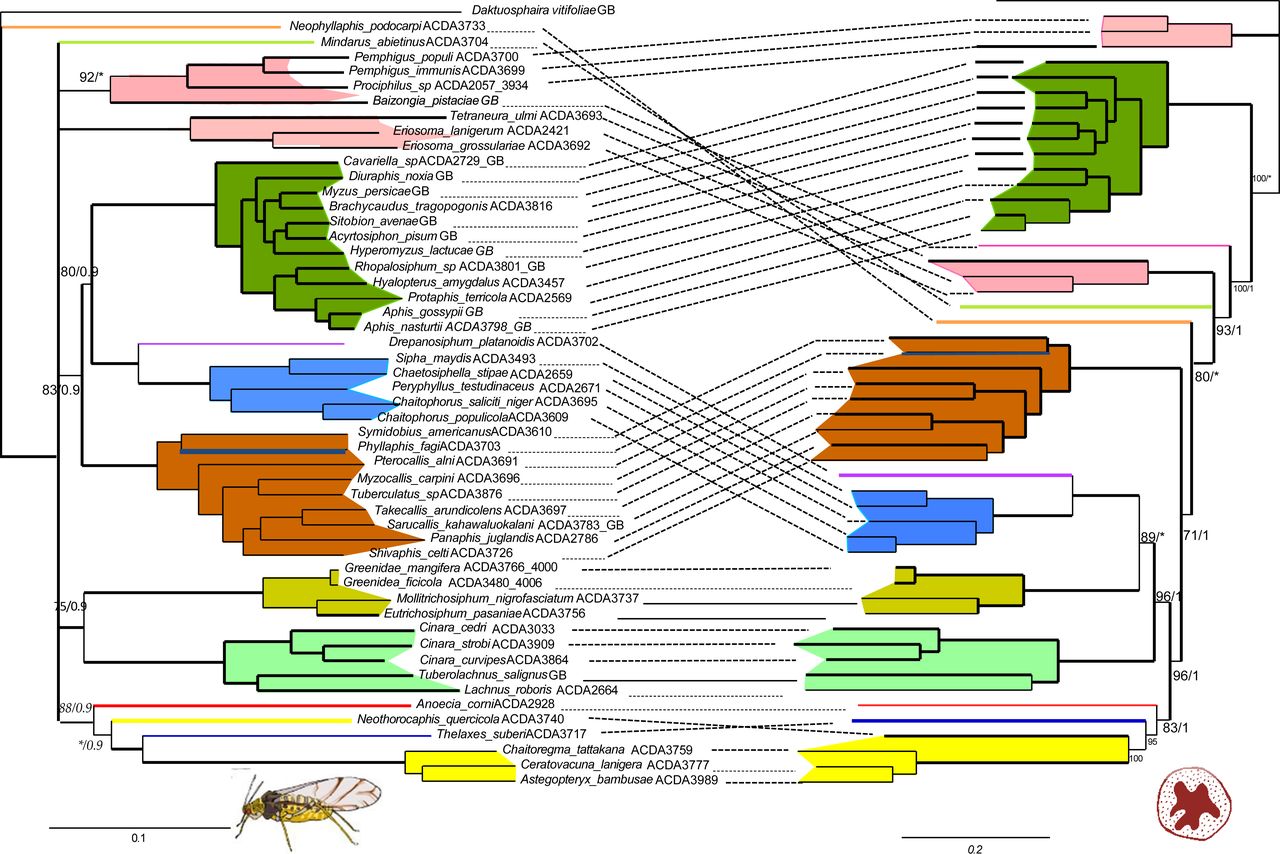
After many years in the making (and an eternal cycle of adding new data….) our work on the discordance among phylogenomic datasets in is out! Relationships...
-
New work out! Serendipitous discovery of a novel leech-associated nutritional symbiont
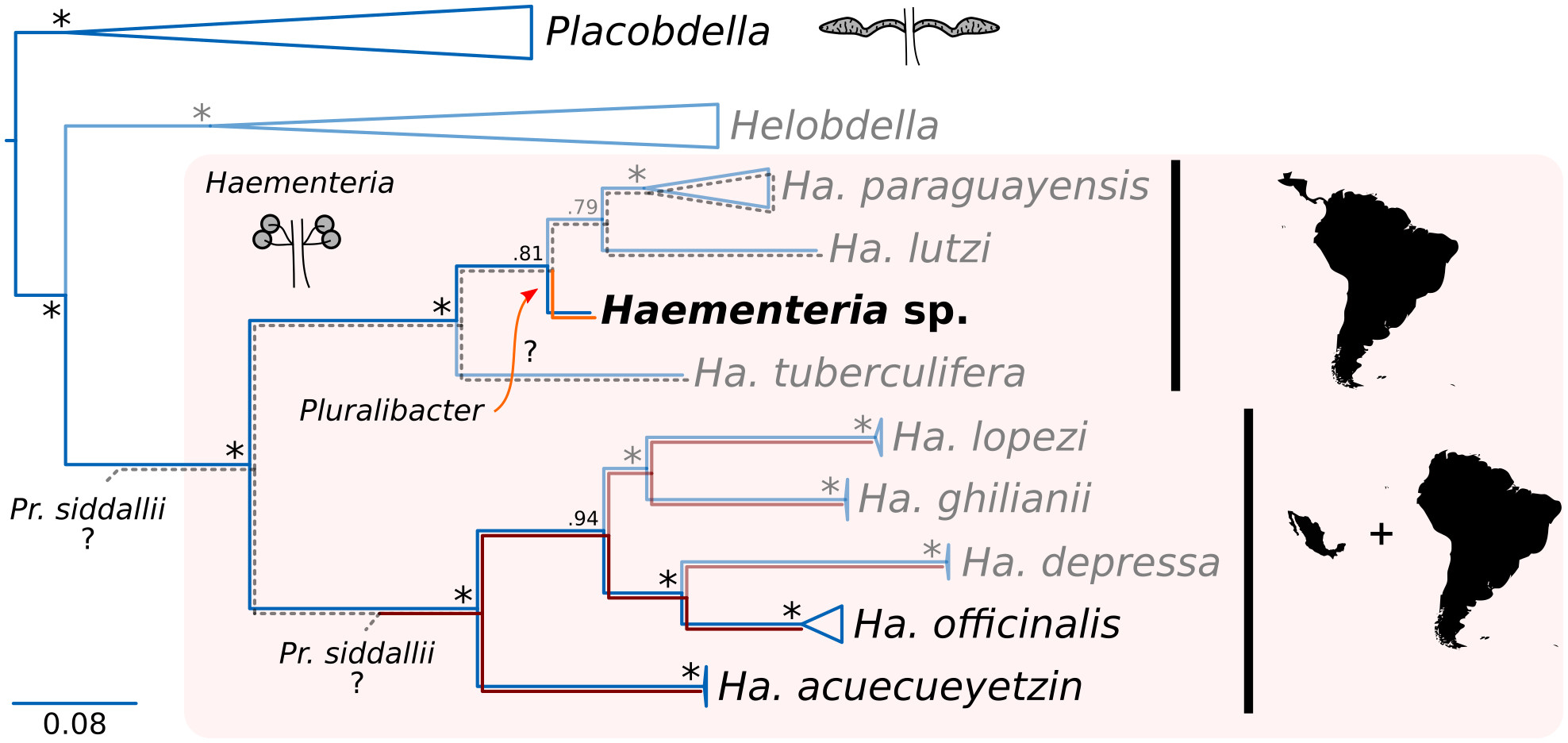
Pluralibacter now joins the (very) selected group of genera from which obligate nutritional symbionts of leeches have evolved from. As the previously known Providencia, this novel...
-
Another great year of the International FISH course at DOME
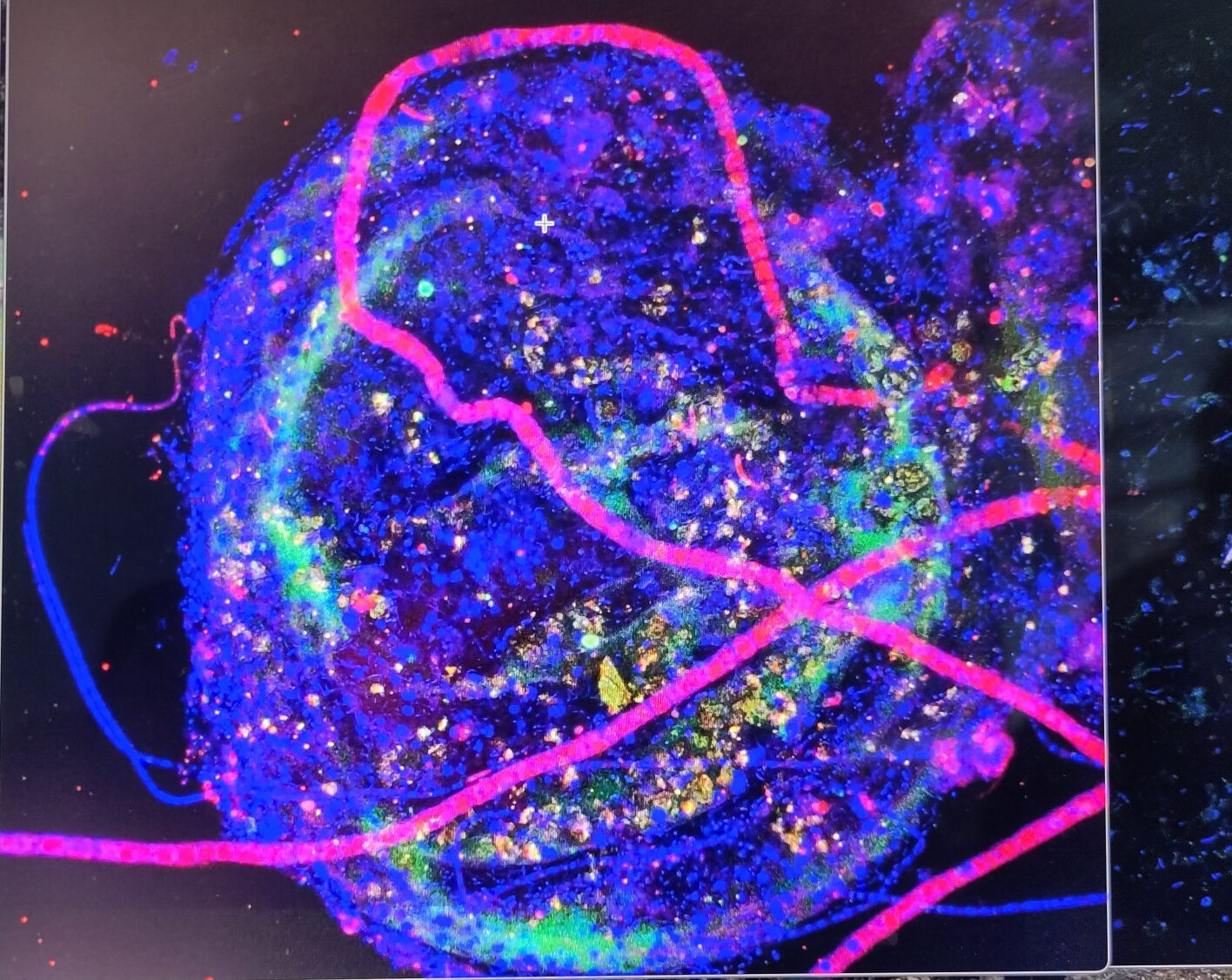
Another year of the International FISH course, where participants bring challenging samples and we help them design and apply the best strategy possible to analyse and...
-
Back from the FEMS 2023 meeting in Hamburg

Back from a great FEMS 2023 edition. This year, a symbiosis session was organised by Martin Kaltenpoth (MPI – Jena). There was a great line-up of...
 Biology and Evolution of Symbiosis (BESymb)
Biology and Evolution of Symbiosis (BESymb)