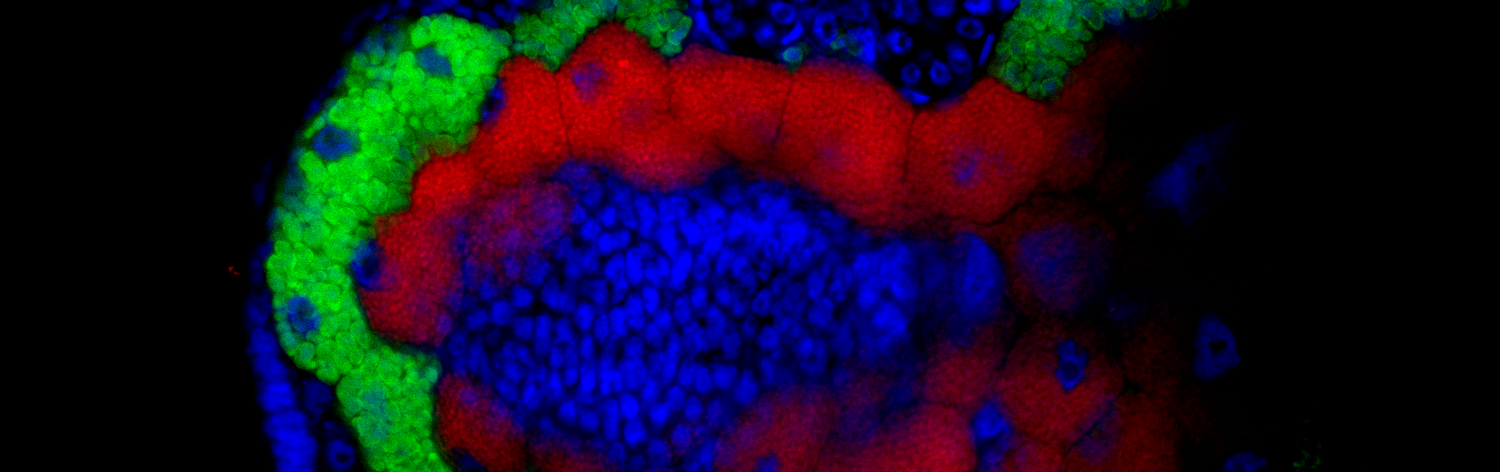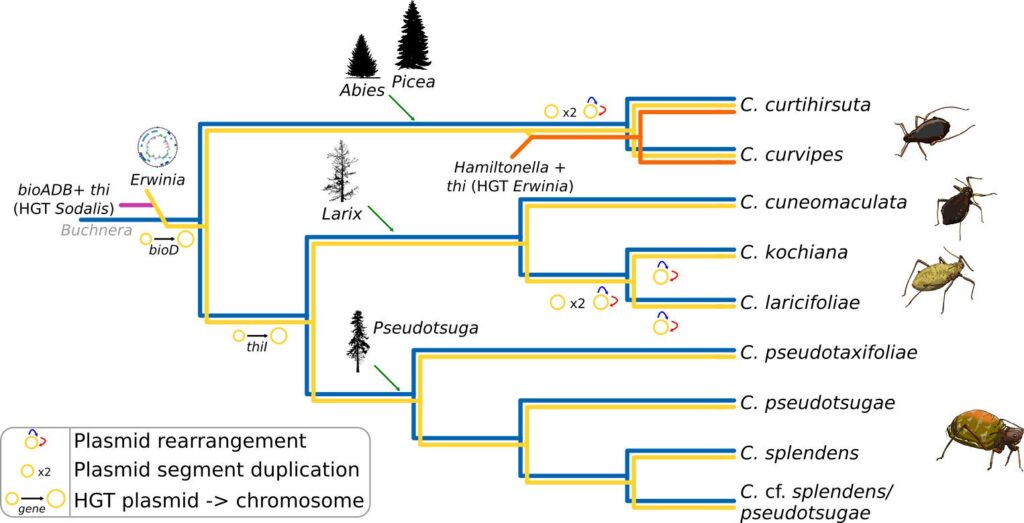
This work was the result of a many many months of exhausting genome assembly, annotation, comparative genomics, and phylogenetics. In this work, we analysed the genome of a newly described co-obligate endosymbiont, Ca. Erwinia haradaeae, of a particular group of aphids within the Cinara genus (Manzano-Marín et. al. 2020). We found that Ca. Erwinia haradaeae, similarly to Buchnera, underwent an early genomic reduction and settled as a co-obligate endosymbiont early before the diversification of the group, likely replacing a pre-existing co-obligate Serratia symbiotica. As expected, they complement Buchnera in key nutritional pathways (riboflavin and biotin) to supply the aphid host with these vitamins. Most significantly, we found these most relevant genes have been horizontally acquired form a Sodalis-like bacterium, highlighting the important role horizontal transfer of genes can play in the establishment and maintenance of beneficial host-microbe associations.
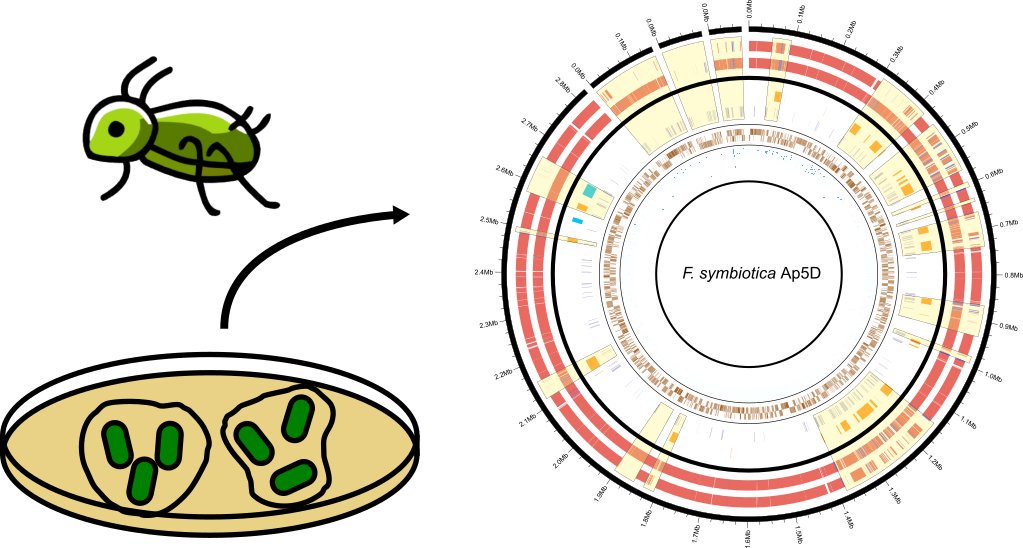
This second work, was the product of a really cool collaboration (I played a very small part) with the Oliver lab and co. (Patel et. al. 2019). Briefly, a facultative Ca. Fukatsuia symbiotica from the pea aphid was cultured in lepidopteran cells and isolated for whole genome sequencing (cool). Full genome analyses and comparative genomics against a co-obligate strain of the same species from a Cinara aphid was performed, and they were not that different. Finally, while Ca. Fukatsuia symbiotica and Ca. Hamiltonella defensa have a strong tendency to co-occur, metabolic complementarity (inferred from available genomes) does not seem to be the basis for this observed characteristic.
 Biology and Evolution of Symbiosis (BESymb)
Biology and Evolution of Symbiosis (BESymb)